Prediction of Lung Cancer Patient Survival via Supervised Machine and Unsupervised Learning Classification Techniques
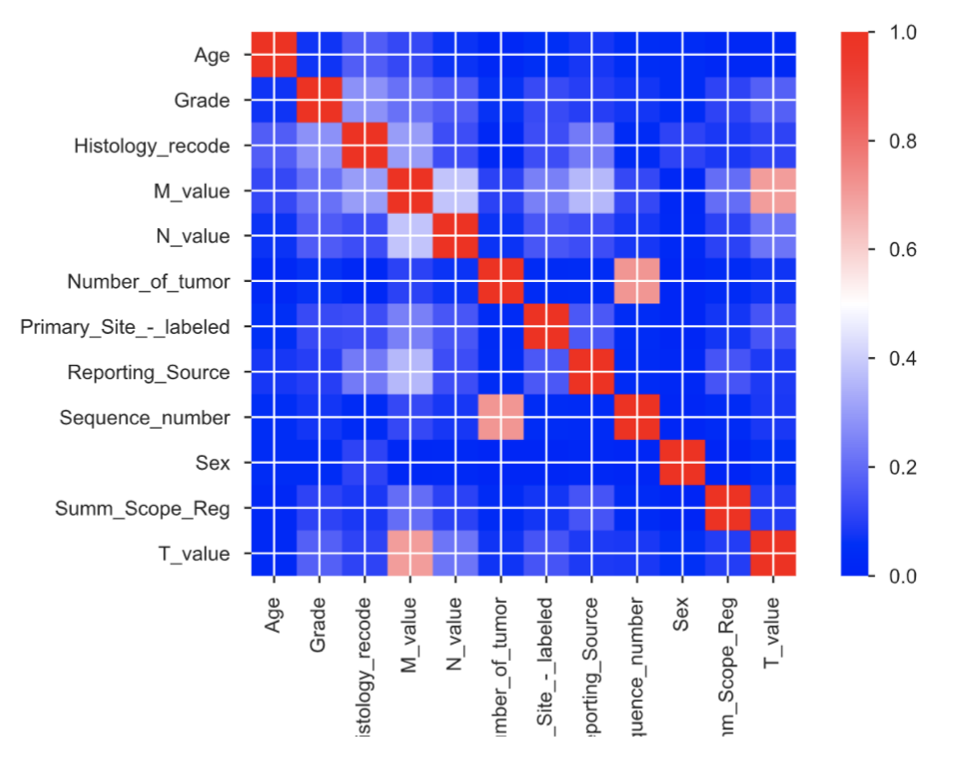
Prediction of Cancer in patients has been estimated by applying various supervised machine learning techniques. These techniques have been applied to a large dataset like Surveillance, Epidemiology, and End Results (SEER) program database. In the past, some researchers have performed different types of classification and regres- sion tasks on the lung cancer datasets but those works did not perform well on large datasets. The major problem in previous research works is the data collection of the patient and the feature selection in the data. The previously proposed supervised models are trained on the data which have no correlation between them. So, the aim of our research project is to build various supervised and unsupervised machine learn- ing models to predict the survival time of lung cancer patients on the given SEER data of lung cancer. And we also perform classification tasks for the prediction of survival time in a different range of years like 0-18 years, 19-30 years. We show an in-depth analysis of various features like tumor-size, age, history, number of lymph nodes values with feature selection techniques like chi2 test, f-regression technique along with the label encoder over different regression and classification technique. The fea- tures have also been selected on the basis correlation value in the correlation matrix. The regression and classification techniques like Linear Regression, SGD Regressor, Random Forest Regressor, Neural network, Customize ensemble have been applied and their performance has been compared by tuning different hyper-parameters. To find the best hyper-parameters we use the cross-validation method. We evaluate the performance of regression models on metrics such as Root Mean Square Error, Stan- dandard Deviation Residual. For classification tasks, we use F1-score, accuracy for evaluation. Out of all models, the neural network model perform exceptionally well on the data having survival month values less than 35 as well as on the data with sur- vival month values less than 72 months. So, we found that semi-supervised learning performs well on this dataset in comparison to supervised learning models.
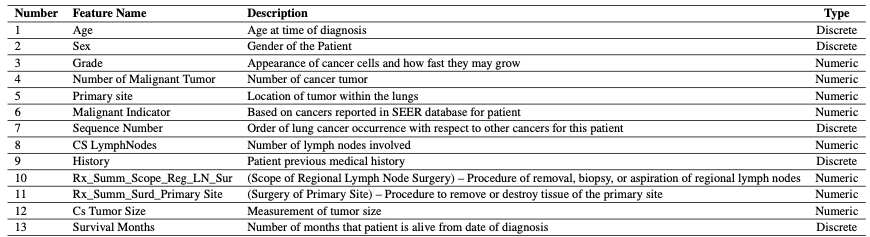
Dataset
Evaluation Parameters:
As mostly we deal with the regression problem so our metric is related to deviaton and error related measures but we also done classification task using the ANN.
-
Root Mean Square Error(RMSE)
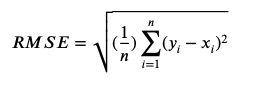
-
Standard Deviation
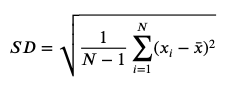
-
Mean
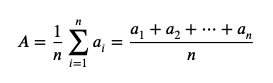
-
Residual Standard Deviation
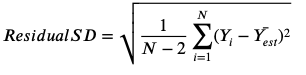
-
Accuracy
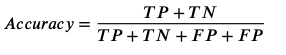
-
F1-score
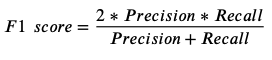
Data Preprocessing:
Change Blank(s) to NaN: In the SEER data, the Null values are represented with keyword Blank(s). But the data frame only handles NaN values so we replace all Blank(s) to NaN value.Handling Null or NaN Values: The SEER which we are using has a high number of null values and NaN values. So first we check the is there any null value in the data. To handle this data we drop those columns which have any null value. So before this step, we have 19 columns in the dataset but after this preprocessing step only 14 columns left in the dataset.Drop rows with the unknown values:The dataset contains columns that have the unknown keywordas value.So we drop those which have unknown as a value in any column. The Grade, Survival months, Number of Malignant tumors having an unknown value in their column. After this step, only 279434 samples remain in the dataset out of 920489 samples.Transform Discrete data to Categorical data: Handling Categorical variables is an important part of preprocessing.The categorical transformation is basically used to convert the discrete and non-continuous data to continuous data. We use ordinal categorical variables with label encoder for converting data into categorical data. So out 13 columns, 8 columns are converted into categorical data.Separation of Survival Data: The survival column is separated from the dataset as it the column for which we have to predict the in our regression model. The survival month column is numeric data which is worked as a final predicted column for our dataset.
Feature Selectiion:
- Univariate linear regression test
- Chi-square test
Models Used:
1. Linear Regression
2. Random Forest Regressor
3. Boosting ensemble technique
4. SGD Regressor
5. Custom Ensemble: Voting Regressor
6. Deep Learning
Results
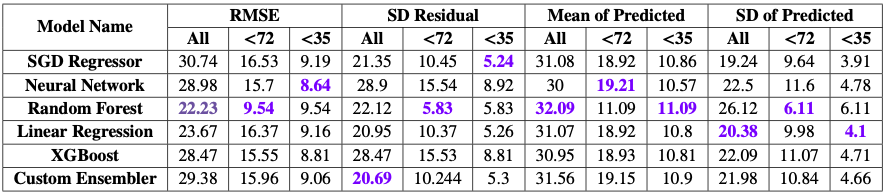
With f-regression test
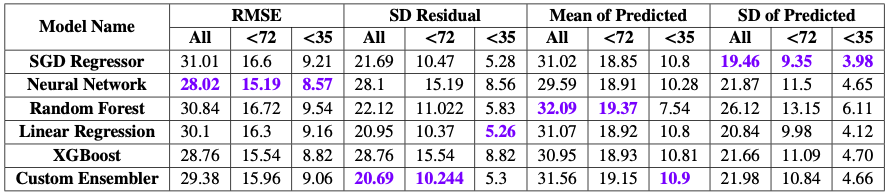
With chi2 test
Contributors:
- Anchit Gupta
- Deekshant Mamodia
- Ankit Agarwal
- Ganesh Chaudhari
All Right Reserved to Repository Owners
MIT License
Copyright (c) 2020 Anchit Gupta
Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the “Software”), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED “AS IS”, WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.